文献汇总
文献汇总
# 生物信息学
config:
target: _blank
imgHeight: 150px
objectFit: cover
lineClamp: 5
data:
- img: https://cdn.ncbi.nlm.nih.gov/pmc/blobs/51c5/10250239/d5c6416f6af9/gkad352figgra1.jpg
link: /pages/90b5a0/
name: ''
desc: <strong>(Jean-Philippe Villemin, Laia Bassaganyas et al. Nucleic Acids Research,2023)</strong> Inferring ligand-receptor cellular networks from bulk and spatial transcriptomic datasets with BulkSignalR
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41592-024-02201-0/MediaObjects/41592_2024_2201_Fig1_HTML.png?as=webp
link: /pages/1c15f5/
name: ''
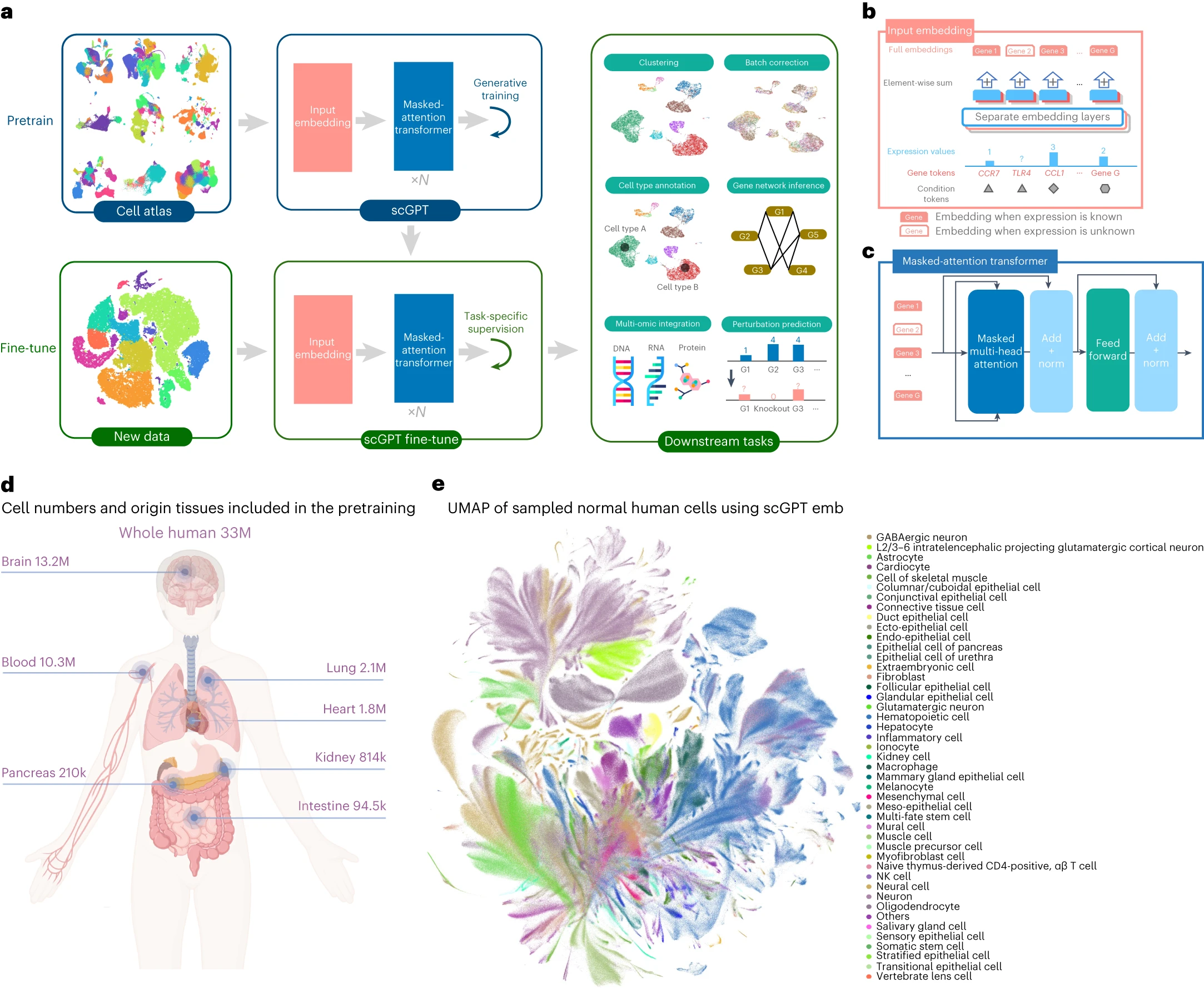
desc: <strong>(Haotian Cui et al. Nat Methods,2024)</strong> scGPT toward building a foundation model for single-cell multi-omics using generative AI
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
# 单细胞空间组
config:
target: _blank
imgHeight: 150px
objectFit: cover
lineClamp: 5
data:
- img: https://genome.cshlp.org/content/31/10/1706/F3.large.jpg
link: /pages/1b45a3/
name: ''
desc: <strong>(Dries R, Chen J et al., Genome Res. 2021)</strong> Advances in spatial transcriptomic data analysis
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41592-022-01480-9/MediaObjects/41592_2022_1480_Fig1_HTML.png?as=webp
link: /pages/e49ae7/
name: ''
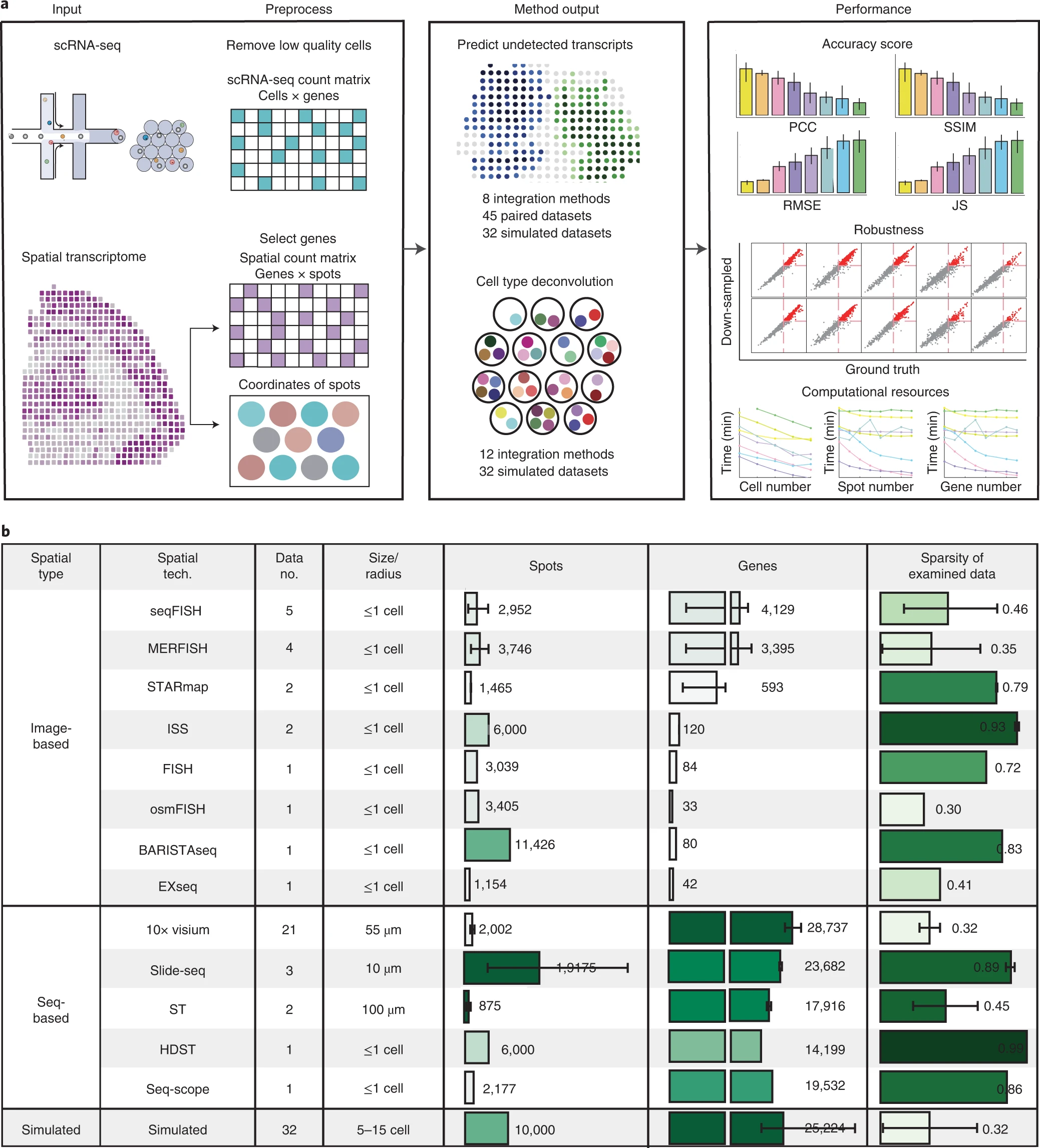
desc: <strong>(Li, B., Zhang, W., Guo, C. et al., Nat Methods ,2022)</strong> Benchmarking spatial and single-cell transcriptomics integration methods for transcript distribution prediction and cell type deconvolution
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41592-023-01773-7/MediaObjects/41592_2023_1773_Fig1_HTML.png?as=webp
link: /pages/d69341/
name: ''
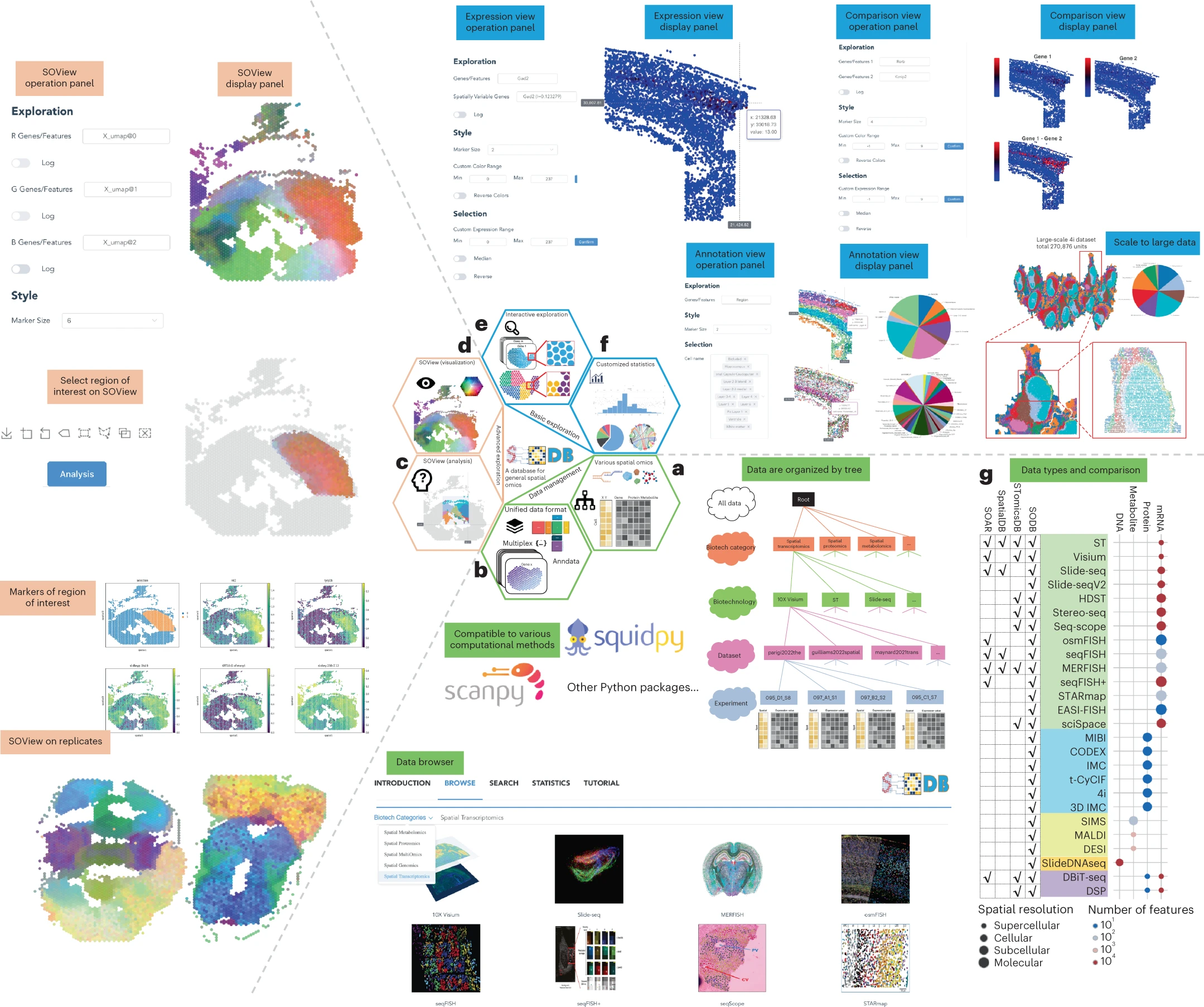
desc: <strong>(Yuan, Z., Pan, W., Zhao, X. et al., Nat Methods,2023)</strong> SODB facilitates comprehensive exploration of spatial omics data
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41587-023-01935-0/MediaObjects/41587_2023_1935_Fig1_HTML.png?as=webp
link: /pages/824737/
name: ''
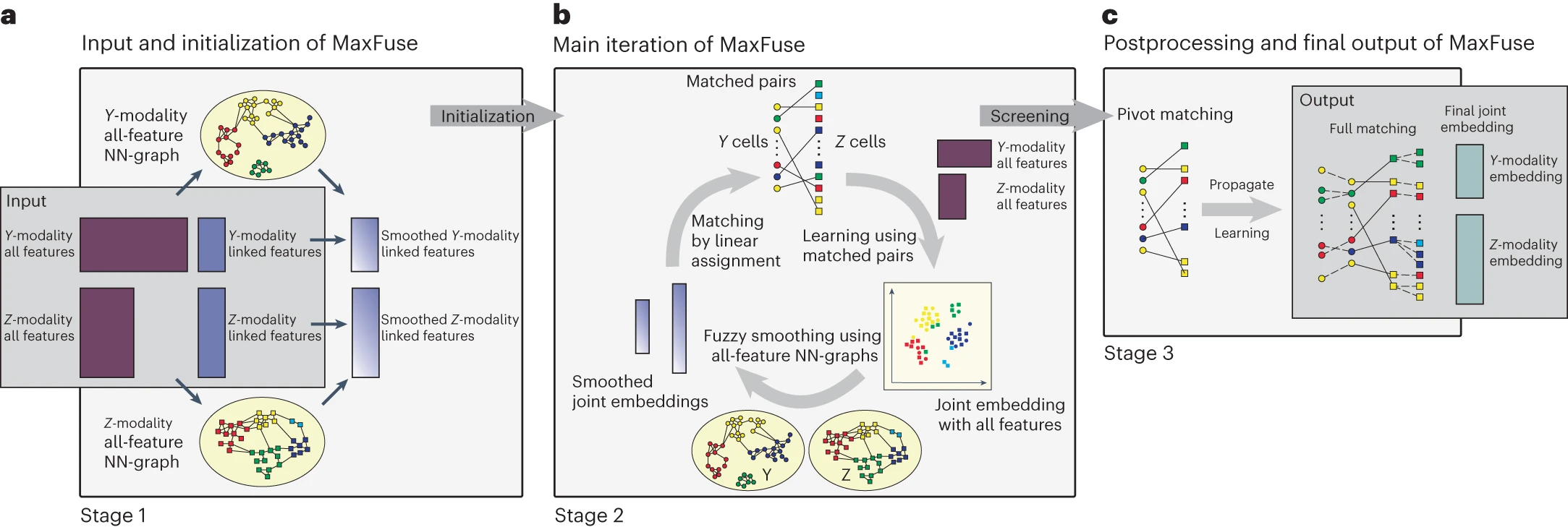
desc: <strong>(Chen, S., Zhu, B., Huang, S. et al., Nat Biotechnol,2023)</strong> Integration of spatial and single-cell data across modalities with weakly linked features
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41592-023-01933-9/MediaObjects/41592_2023_1933_Fig1_HTML.png?as=webp
link: /pages/c733df/
name: ''
desc: <strong>(Grabski, I.N., Street, K. & Irizarry, R.A., Nat Methods,2023)</strong> Significance analysis for clustering with single-cell RNA-sequencing data
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41586-023-06812-z/MediaObjects/41586_2023_6812_Fig1_HTML.png?as=webp
link: /pages/58ba19/
name: ''
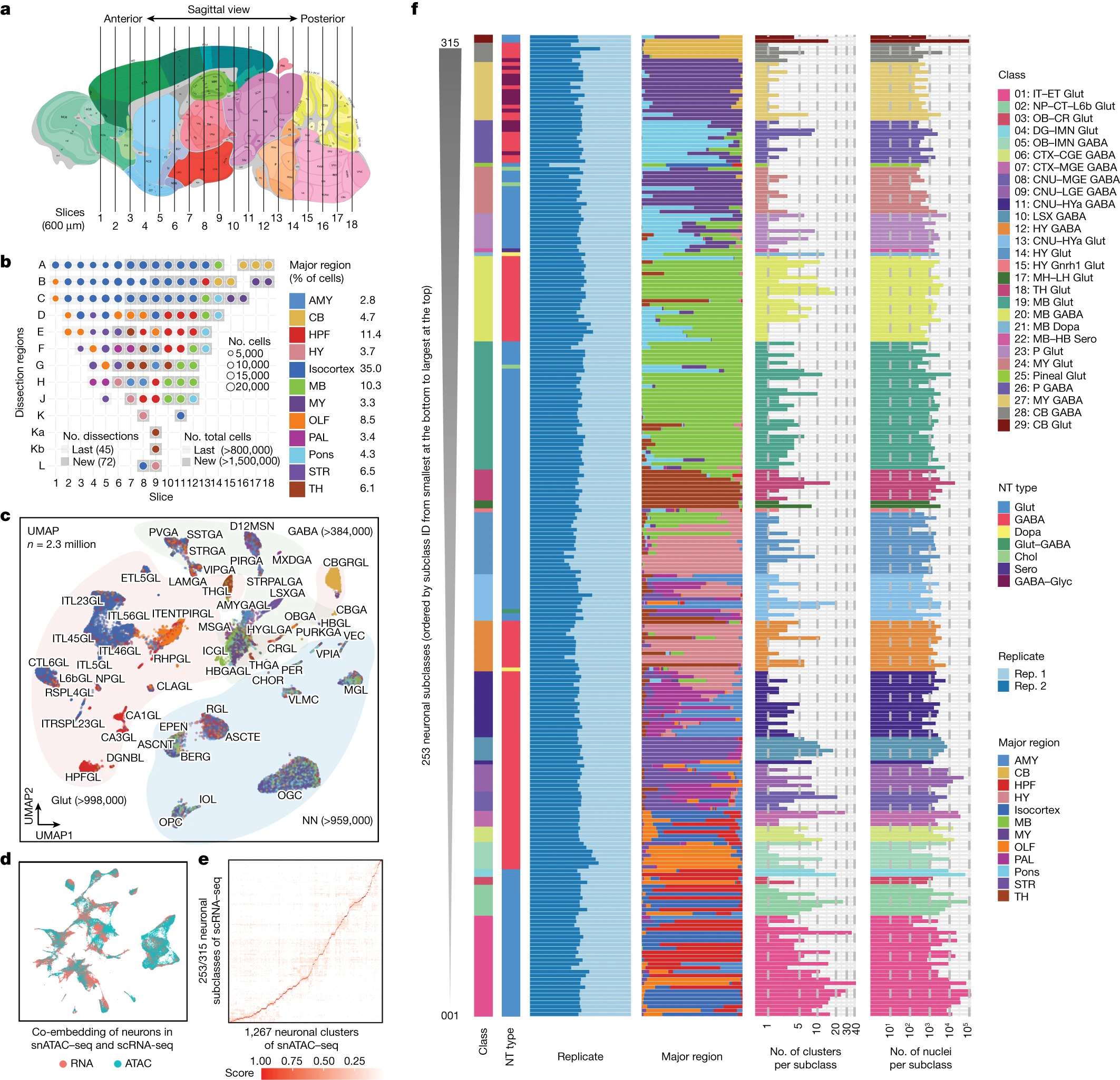
desc: <strong>(Yao, Z., van Velthoven, C.T.J., Kunst, M. et al., Nature,2023)</strong> A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41592-024-02215-8/MediaObjects/41592_2024_2215_Fig1_HTML.png?as=webp
link: /pages/7065f3/
name: ''
desc: <strong>(Yuan, Z., Zhao, F., Lin, S. et al., Nat Methods, 2024)</strong> Benchmarking spatial clustering methods with spatially resolved transcriptomics data
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
# 免疫学相关
config:
target: _blank
imgHeight: 150px
objectFit: cover
lineClamp: 5
data:
- img: https://www.cell.com/cms/attachment/cc0d6647-7430-41ee-8bea-fe63f95f2d22/fx1.jpg
link: /pages/417e50/
name: ''
desc: <strong>(Fei Tang,Jinhu Li,Lu Qi et al. Cell,2023)</strong> A pan-cancer single-cell panorama of human natural killer cells
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41467-023-42814-1/MediaObjects/41467_2023_42814_Fig1_HTML.png?as=webp
link: /pages/0f3668/
name: ''
desc: <strong>(Shi, DD., Zhang, YD., Zhang, S. et al. Nat Commun,2023)</strong> Stress-induced red nucleus attenuation induces anxiety-like behavior and lymph node CCL5 secretion
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41586-023-06816-9/MediaObjects/41586_2023_6816_Fig1_HTML.png?as=webp
link: /pages/c3a7f3/
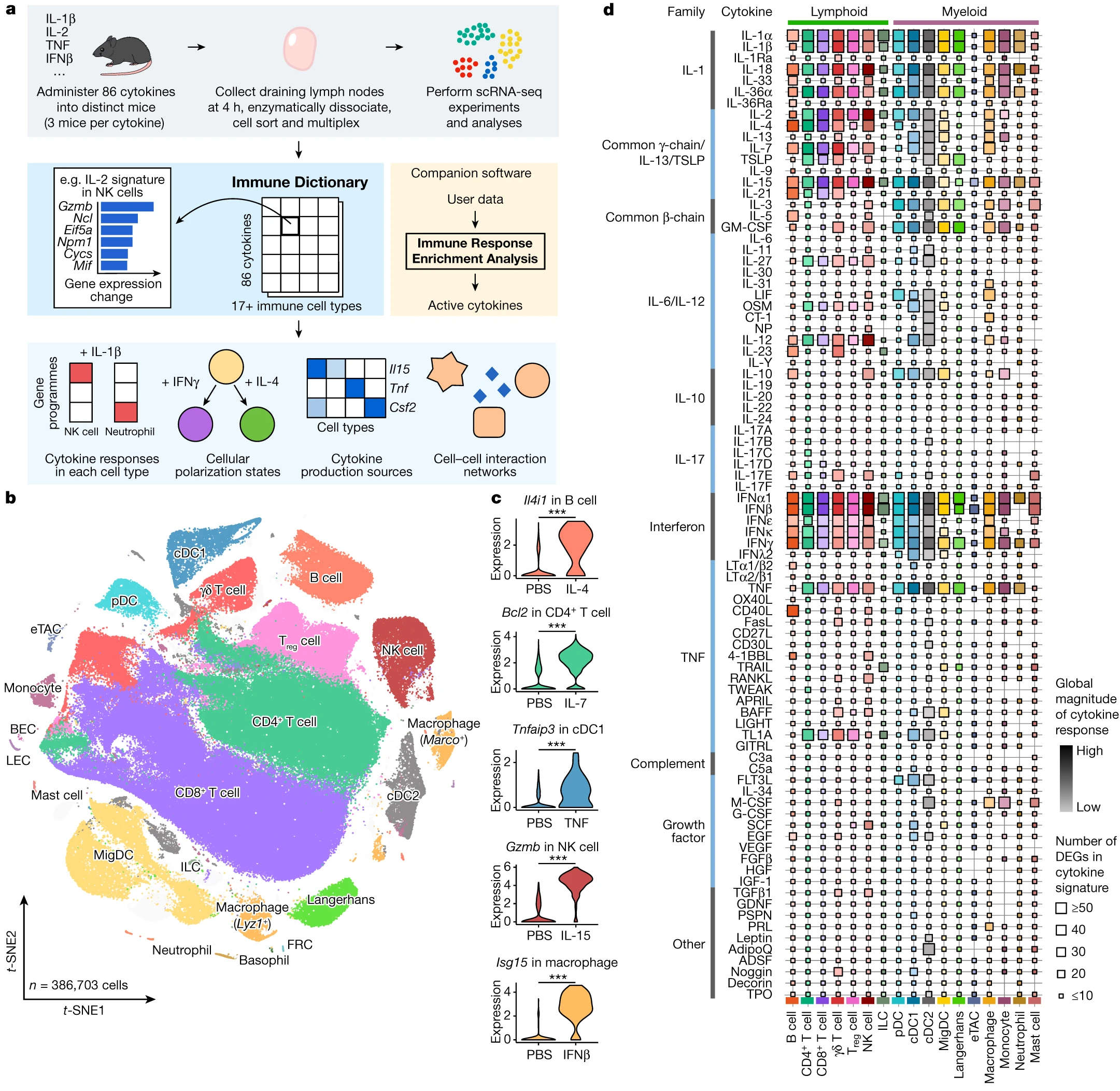
name: ''
desc: <strong>(Cui, A., Huang, T., Li, S. et al. Nature,2023)</strong> Dictionary of immune responses to cytokines at single-cell resolution
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
# scRNA-scATAC整合
config:
target: _blank
imgHeight: 150px
objectFit: cover
lineClamp: 5
data:
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fnmeth.4401/MediaObjects/41592_2017_BFnmeth4401_Fig1_HTML.jpg?as=webp
link: /pages/c6e85a/
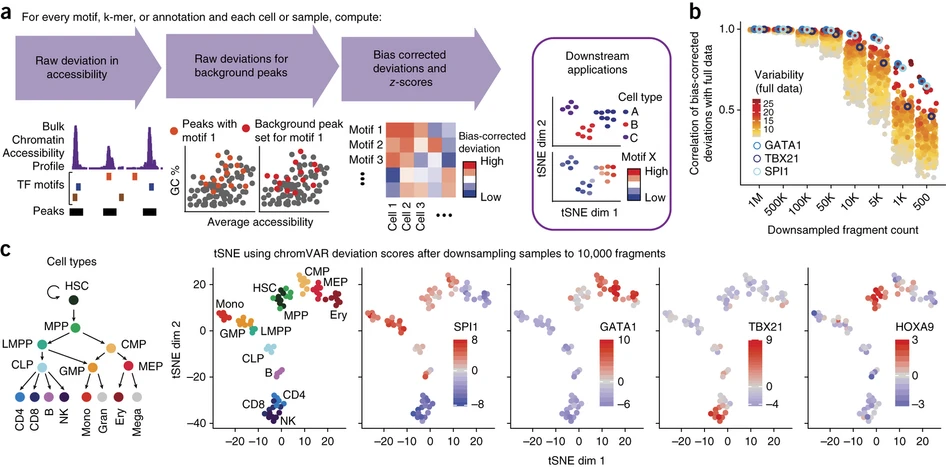
name: ''
desc: <strong>(Schep, A., Wu, B., Buenrostro, J. et al. Nat Methods,2017)</strong> chromVAR inferring transcription-factor-associated accessibility from single-cell epigenomic data
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41467-022-34413-3/MediaObjects/41467_2022_34413_Fig1_HTML.png?as=webp
link: /pages/7ecadd/
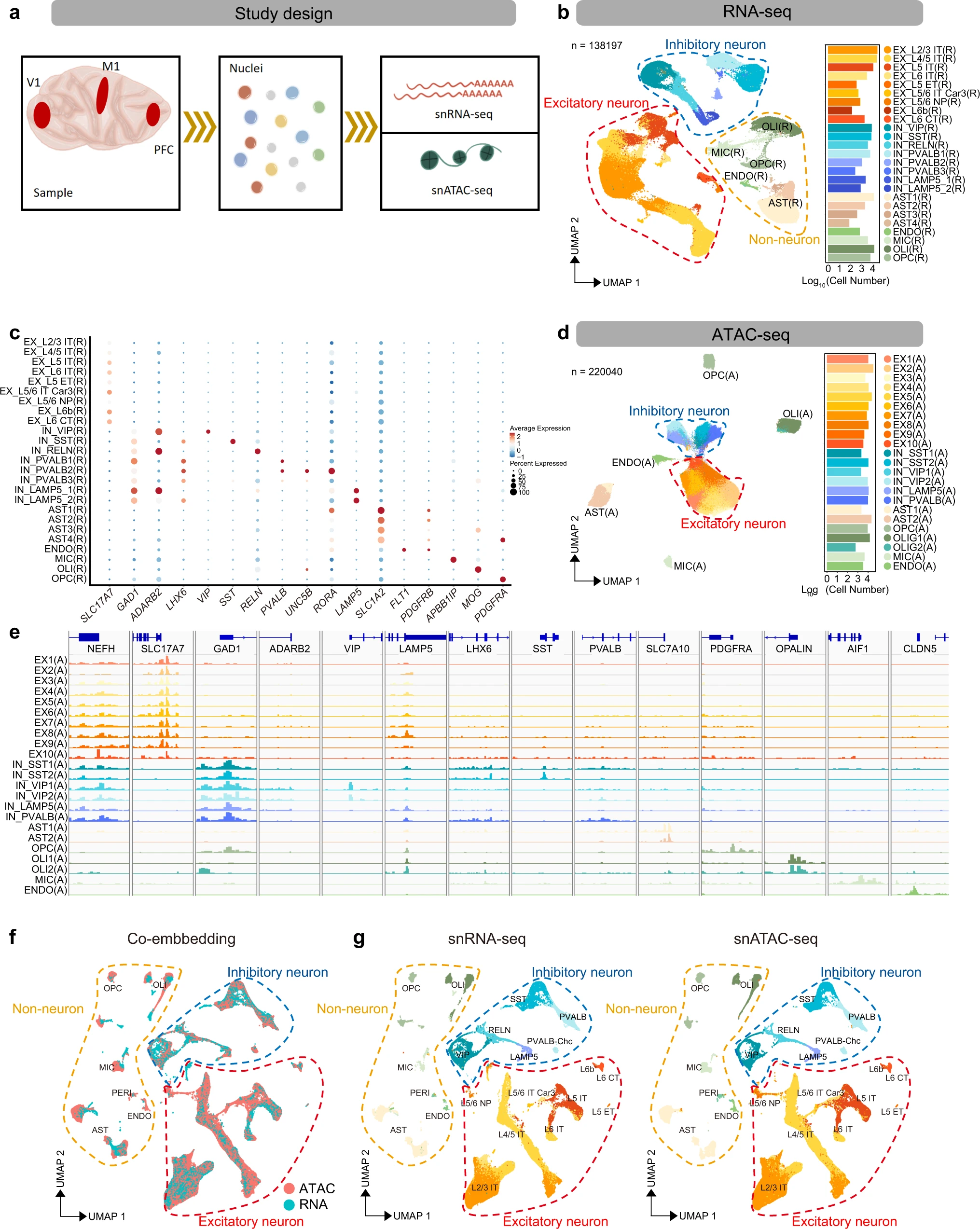
name: ''
desc: <strong>(Lei Y, Cheng M et al. Nat Commun,2022)</strong> Spatially resolved gene regulatory and disease-related vulnerability map of the adult Macaque cortex
- img: https://www.cell.com/cms/attachment/c70e347b-3921-45c3-be57-e455c91976bc/gr1.jpg
link: /pages/41e09e/
name: ''
desc: <strong>(Zhongli Xu, Xinjun Wang et al. iScience,2022)</strong> Integrative analysis of spatial transcriptome with single-cell transcriptome and single-cell epigenome in mouse lungs after immunization
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41586-022-05094-1/MediaObjects/41586_2022_5094_Fig1_HTML.png?as=webp
link: /pages/587589/
name: ''
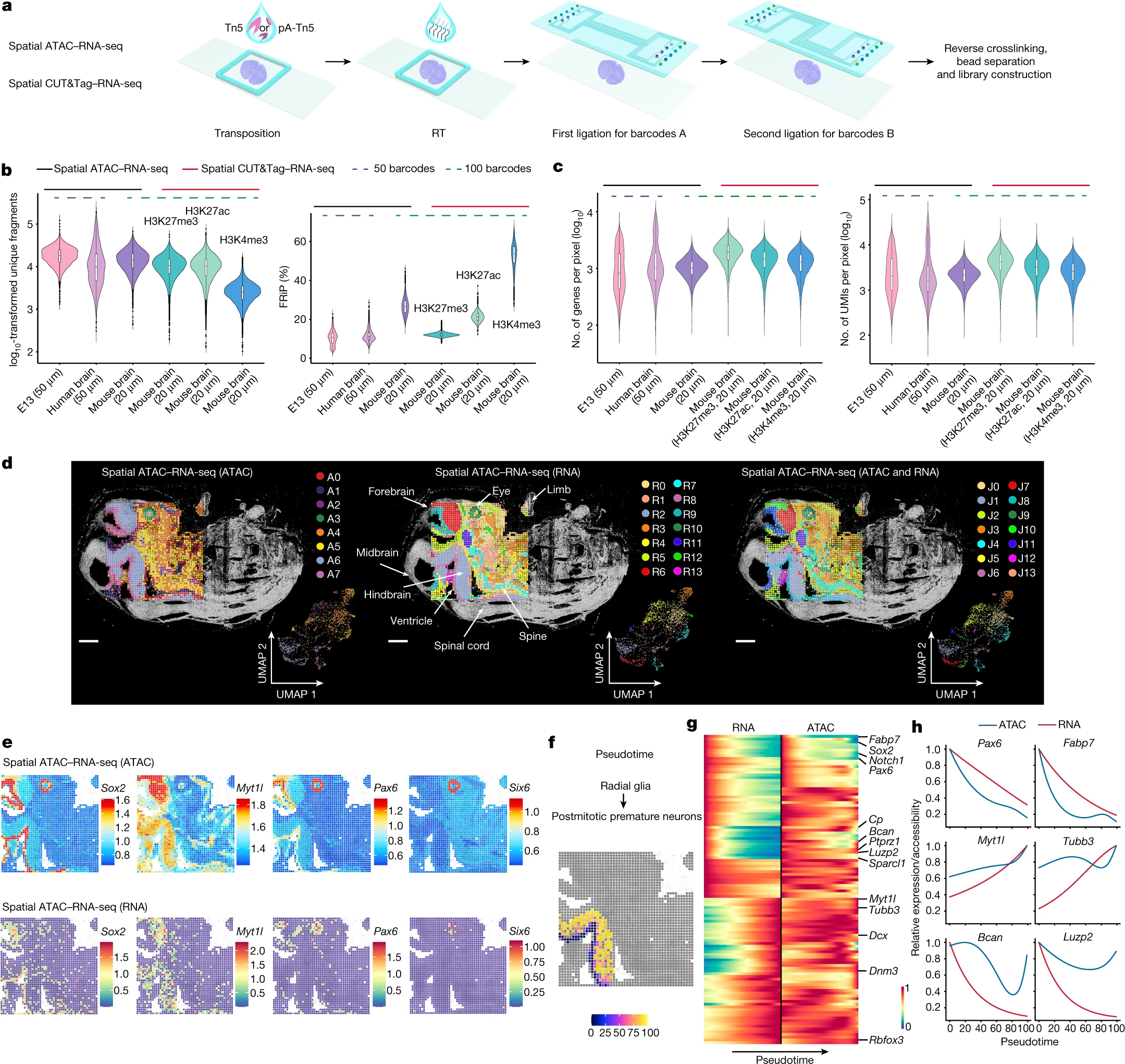
desc: <strong>(Deng, Y., Bartosovic, M., Ma, S. et al. Nature,2022)</strong> Spatial profiling of chromatin accessibility in mouse and human tissues
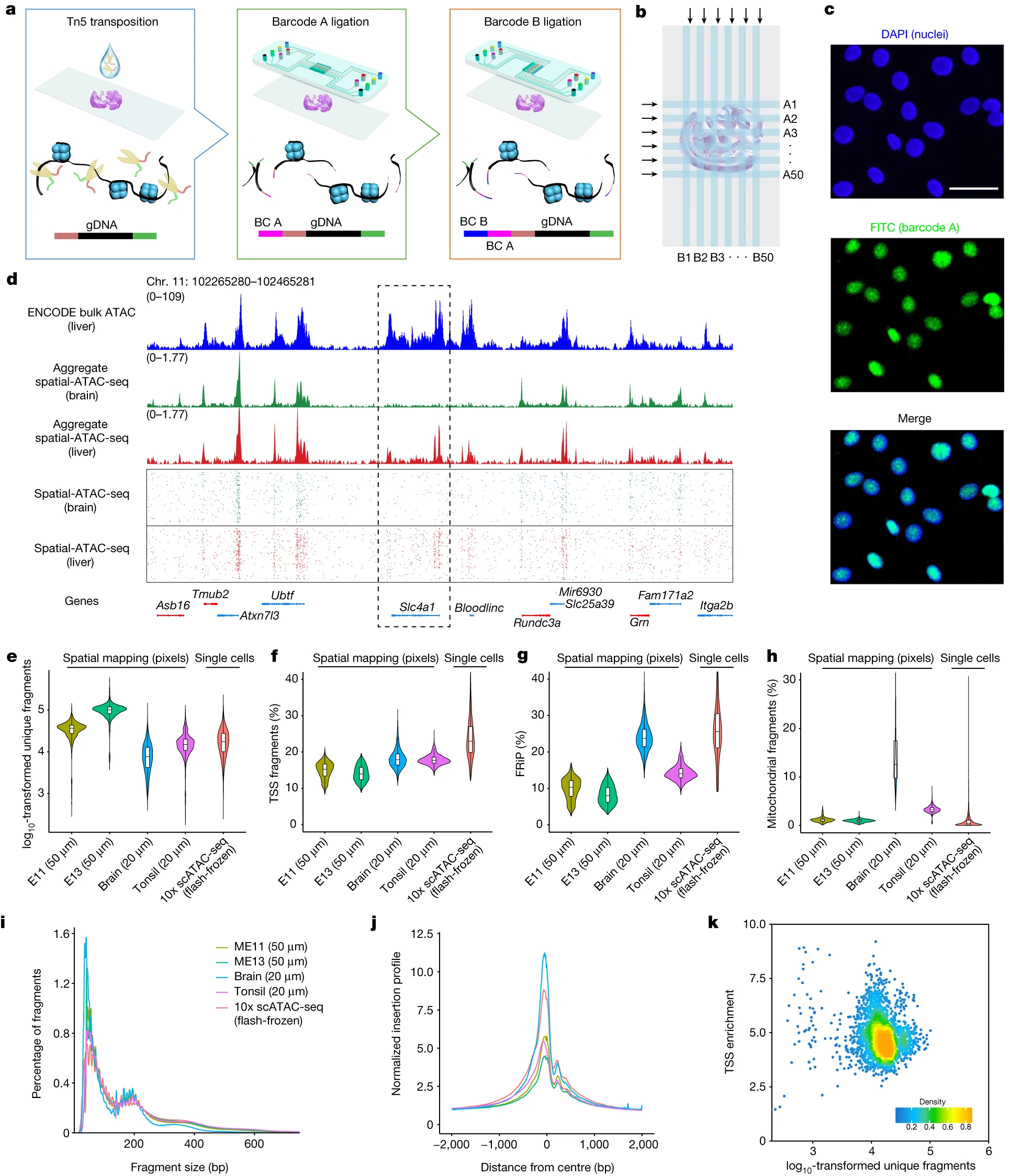
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41586-023-05795-1/MediaObjects/41586_2023_5795_Fig1_HTML.png?as=webp
link: /pages/9abd5e/
name: ''
desc: <strong>(Zhang, D., Deng, Y., Kukanja, P. et al. Nature,2023)</strong> Spatial epigenome-transcriptome co-profiling of mammalian tissues
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41586-023-06824-9/MediaObjects/41586_2023_6824_Fig1_HTML.png?as=webp
link: /pages/fa0e3f/
name: ''
desc: <strong>(Zu, S., Li, Y.E., Wang, K. et al. Nature,2023)</strong> Single-cell analysis of chromatin accessibility in the adult mouse brain
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41586-023-06682-5/MediaObjects/41586_2023_6682_Fig1_HTML.png?as=webp
link: /pages/17d203/
name: ''
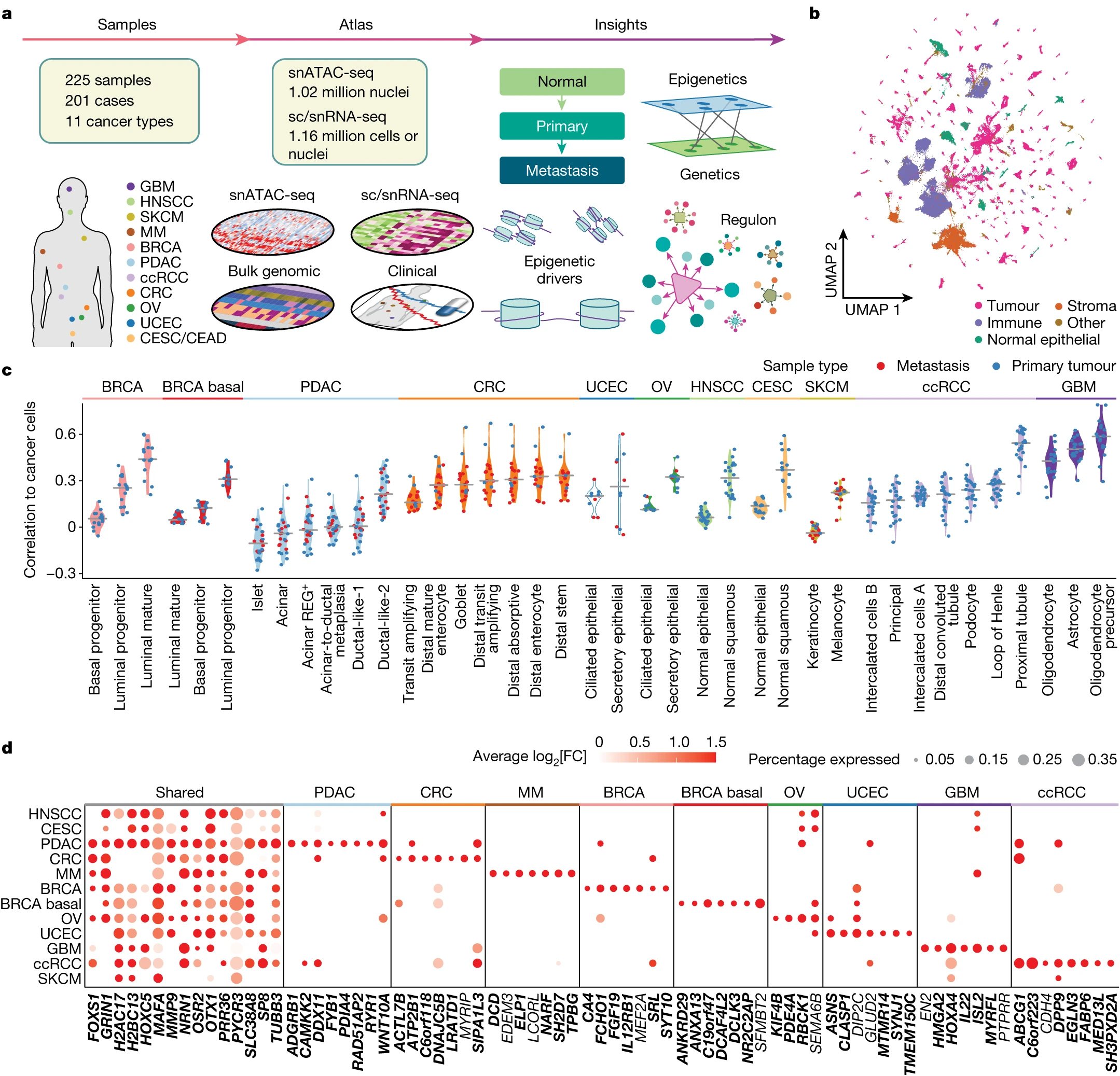
desc: <strong>(Terekhanova, N.V., Karpova, A., Liang, WW. et al. Nature,2023)</strong> Epigenetic regulation during cancer transitions across 11 tumour types
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
# 计算生物学
config:
target: _blank
imgHeight: 150px
objectFit: cover
lineClamp: 5
data:
- img: https://media.springernature.com/full/springer-static/image/art%3A10.1038%2Fs41467-025-65362-2/MediaObjects/41467_2025_65362_Fig1_HTML.png?as=webp
link: /pages/8ebfe8/
name: ''
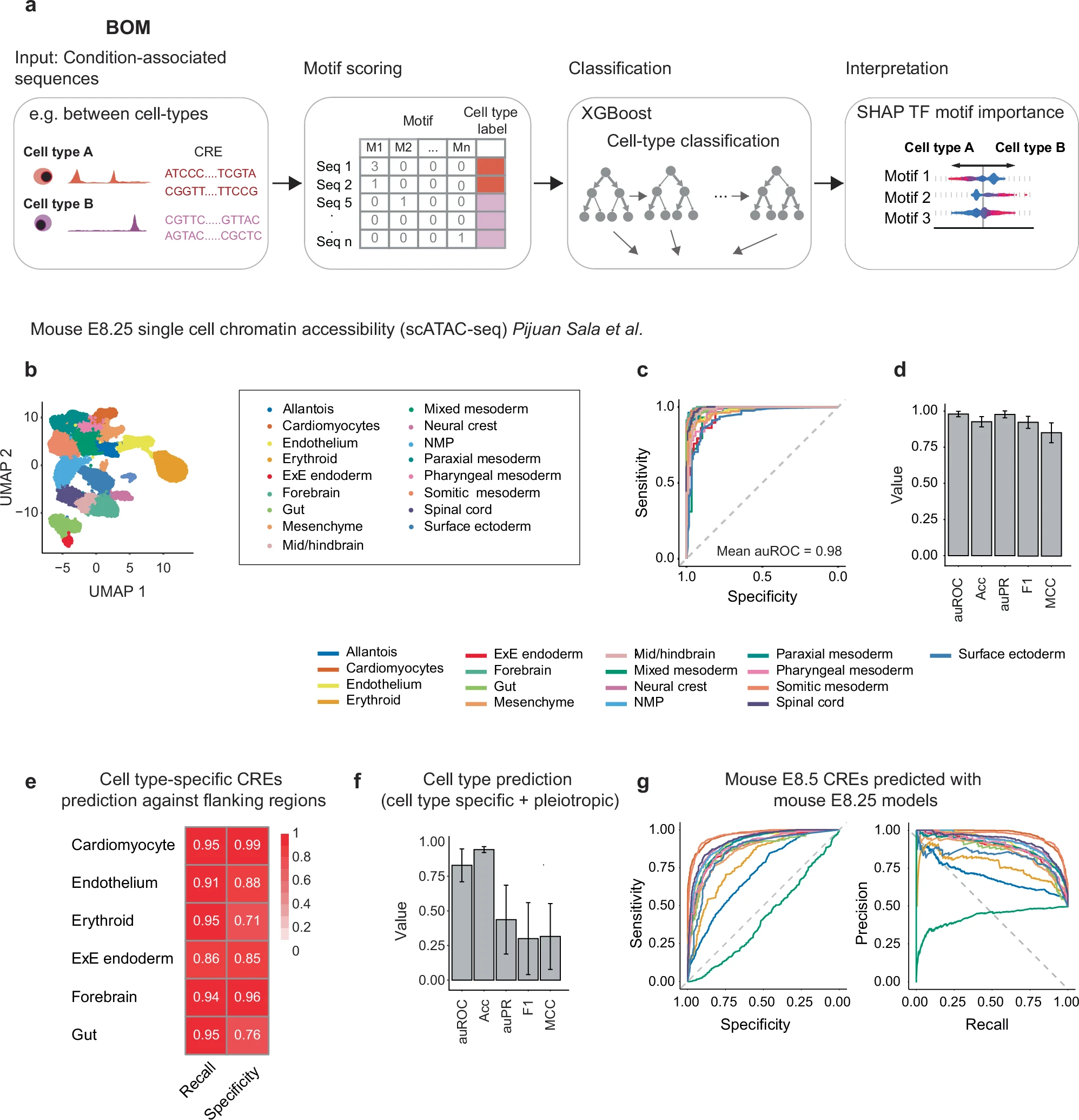
desc: <strong>(Cornejo-Páramo, P., Zhang, X., Louis, L. et al. Nat Commun,2025)</strong> Motif-based models accurately predict cell type-specific distal regulatory elements
1
2
3
4
5
6
7
8
9
10
11
12
2
3
4
5
6
7
8
9
10
11
12
编辑 (opens new window)